Tissue Registration
WSI.tissue_registration module
About this module
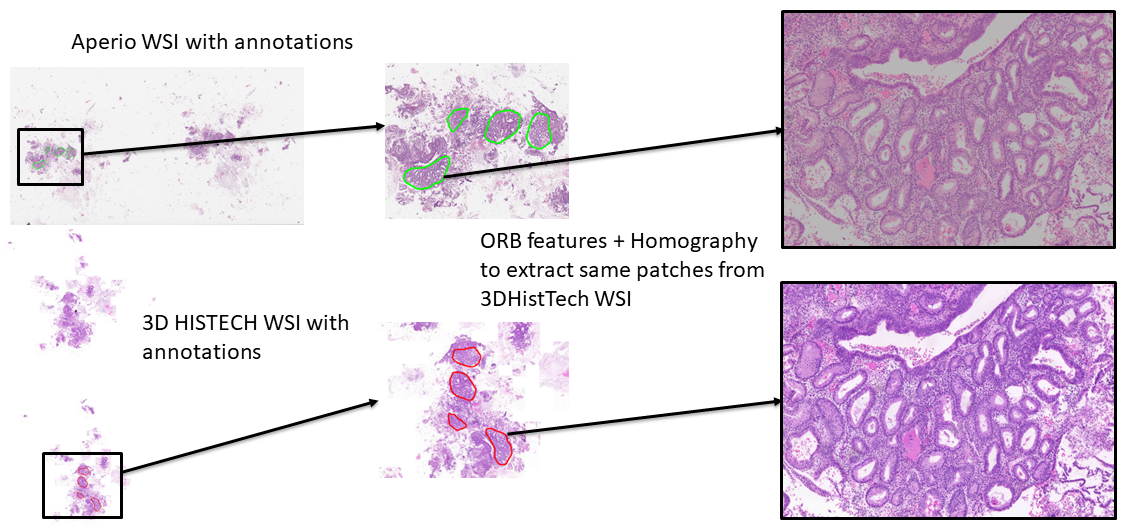
This module enables tissue registration and annotation transfer between Whole Slide Images (WSIs) scanned by different devices (e.g., Aperio and Histech). It uses ORB-based image registration and homography mapping to align annotations and extract corresponding regions.
The key class is tissue_registration, which contains all logic to perform: - ORB-based alignment - Annotation transformation via XML parsing - Patch extraction - Registration visualization
Loading Required Packages
from WSI.tissue_registration import tissue_registration
You’ll need to install the following packages:
pip install openslide-python tiffslide lxml opencv-python-headless
Input Structure
To use the tissue_registration module, prepare the following files:
Aperio Whole Slide Image (WSI): typically a .svs file from Aperio scanner.
Histech Whole Slide Image (WSI): typically a .svs file from Histech scanner.
Aperio XML annotation file: contains region-level annotations that need to be transferred to Histech image space.
These inputs are used to align slides from different scanners and propagate annotations across them.
Define the paths and output directory:
from registration import tissue_registration
reg = tissue_registration(
aperio_path="slides/Aperio1.svs",
histech_path="slides/Histech1.svs",
xml_path="annotations/Aperio1.xml",
output_base="output/registration/"
)
Make sure that the output_base directory exists or will be created, as it is used to store visual results and annotation outputs.
Running the Pipeline
To perform the full registration and annotation transfer process, use the .run() method:
reg.run()
This method executes the following sequence:
Load WSIs and extract level-2 thumbnails for faster processing.
Detect ORB keypoints and compute descriptors in both Aperio and Histech images.
Estimate homography to map Aperio coordinates into Histech space.
Parse Aperio XML annotation file to get annotated regions.
Transform annotations to match the coordinate system of Histech WSI.
Extract tissue patches around the transformed annotations.
Generate visual overlays showing registration alignment and transferred annotations.
Save updated XML containing mapped annotations compatible with Histech image.
Visualizing the Results
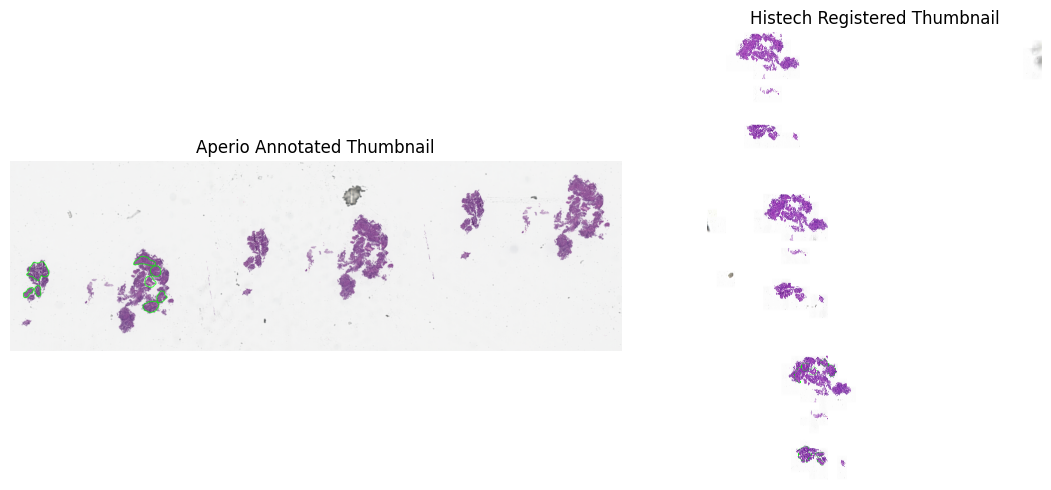
After running the pipeline, you can visualize the thumbnails with overlaid annotations:
reg.show_annotated_thumbnails()
To view paired extracted patches side by side:
reg.show_registered_patches(max_pairs=2)

Outputs
After running .run(), your output_base directory will contain:
/aperio_patches: Aligned patches extracted from Aperio WSI
/histech_patches: Corresponding patches extracted from Histech WSI
/visualizations: Annotated thumbnails and overlays
—
### Notes: - Ensure all WSIs and XML annotations are correctly paired and accessible. - Level 2 thumbnails are used for registration to speed up ORB computation.