Duplicate Detection
WSI.hashing module
About this module
This documentation page describes the duplicate-detection workflow used for Whole Slide Images (WSIs).
Duplicate detection compares two WSIs by: 1. Generating perceptual hashes for each slide 2. Computing the Normalized Hamming Distance between the hashes
A lower normalized Hamming distance indicates higher similarity (possible duplicates), while a higher value indicates greater dissimilarity.
The core API used is:
WSI.hashing.calculate_hamming_distance(first_path, second_path, rotation=..., resizepercentage=...)
Duplicate Detection Results
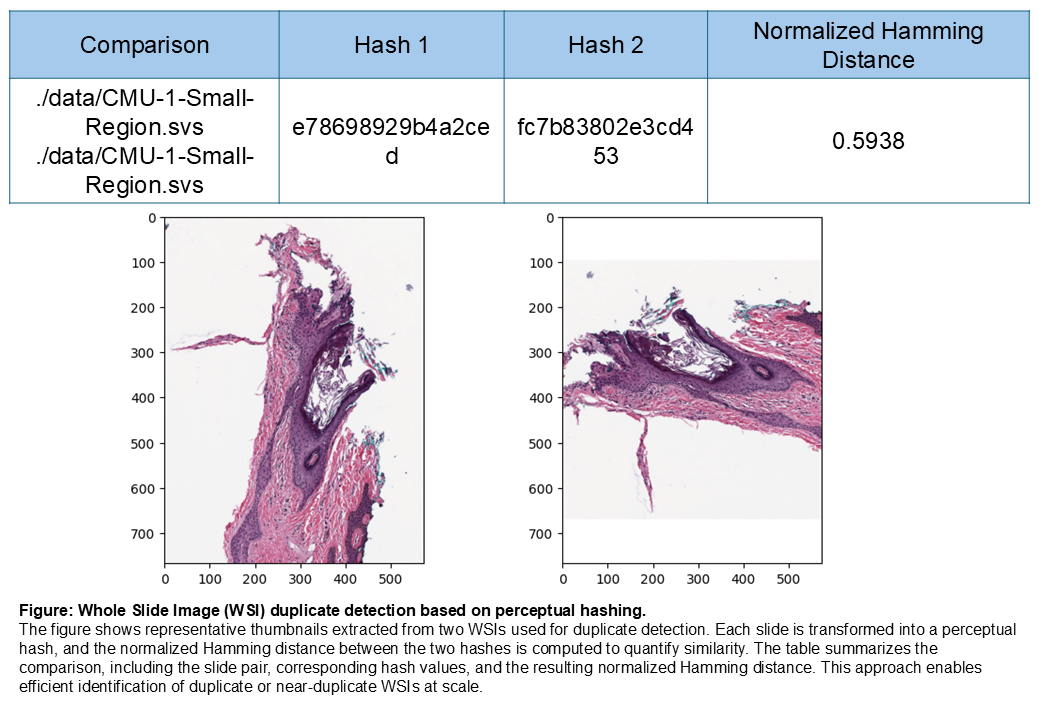
Comparison |
Hash 1 |
Hash 2 |
Normalized Hamming Distance |
|---|---|---|---|
./data/CMU-1-Small-Region.svs - ./data/CMU-1-Small-Region.svs |
e78698929b4a2ced |
fc7b83802e3cd453 |
0.5938 |
Figure: Duplicate detection example
The figure below shows thumbnails from the two WSIs used for comparison and the resulting perceptual hashes with the computed normalized Hamming distance.
Loading Required Packages
import os
import matplotlib.pyplot as plt
from WSI.readwsi import WSIReader
from WSI.hashing import calculate_hamming_distance
Compute Normalized Hamming Distance
Use calculate_hamming_distance to compute the normalized Hamming distance between two WSIs.
Parameters commonly used:
- rotation: rotate the thumbnail/representation before hashing (degrees)
- resizepercentage: percent scale used before hashing (e.g., 100, 10, 0.5)
first_path = "./data/CMU-1-Small-Region.svs"
second_path = "./data/CMU-1-Small-Region.svs"
calculate_hamming_distance(first_path, second_path, rotation=0, resizepercentage=10)
Interpreting Results
Lower normalized distance -> slides are more similar (possible duplicates)
Higher normalized distance -> slides are less similar
You can define a duplicate threshold based on your dataset and scanner/staining variation.
Usage Example (Minimal)
from WSI.hashing import calculate_hamming_distance
first_path = "./data/slide_A.svs"
second_path = "./data/slide_B.svs"
calculate_hamming_distance(first_path, second_path, rotation=0, resizepercentage=10)