About DomId¶
VaDE Model Summary¶
Variational Deep Embedding (VaDE) [1] is an unsupervised generative clustering approach based on Variational Autoencoders [2].
In this library, VaDE can be deployed as a deep clustering model using both Artificial Neural Network (ANN) and Convolutional Neural Network (CNN) architectures for the encoder (x,φ) and the decoder f(z,θ).
The encoder learns to compress the high-dimensional input images x into lower-dimensional latent representations z.
Using a Mixture-of-Gaussians (MOG) prior distribution for the latent representations z, we examine sub-groups or domains within the dataset, revealed by the individual Gaussians within the learned latent space, and how z affects the generation of x.
The model can be used to perform inference, where observed images x are mapped to a series of corresponding latent variables z and their cluster/domain assignments c.
We denote the latent space dimensionality by d (i.e., z ∈ R^d), and the number of clusters by D (i.e., c ∈ {1, 2, …, D}).
The decoder CNN generates images x from latent space samples z. Thus, the trained decoder CNN can also be used to generate synthetic images from the algorithmically identified subgroups. VaDE is optimized using Stochastic Gradient Variational Bayes cite{kingmaAutoEncodingVariationalBayes2013} to maximize a statistical measure called the Evidence Lower Bound (ELBO).
CDVaDE Model Summary¶
In this package, we also implement the Conditionally Decoded Variational Deep Embedding (CDVaDE) model [4] as an expansion to VaDE. CDVaDE’s generative process is different from VaDE’s because it combines extra variables y with the latent representation z, as shown in the Figure below. These variables may include class labels or existing subgroup structures that do not need to be identified by the clustering algorithm. It is expected that these additional variables y are accessible during both training and testing.
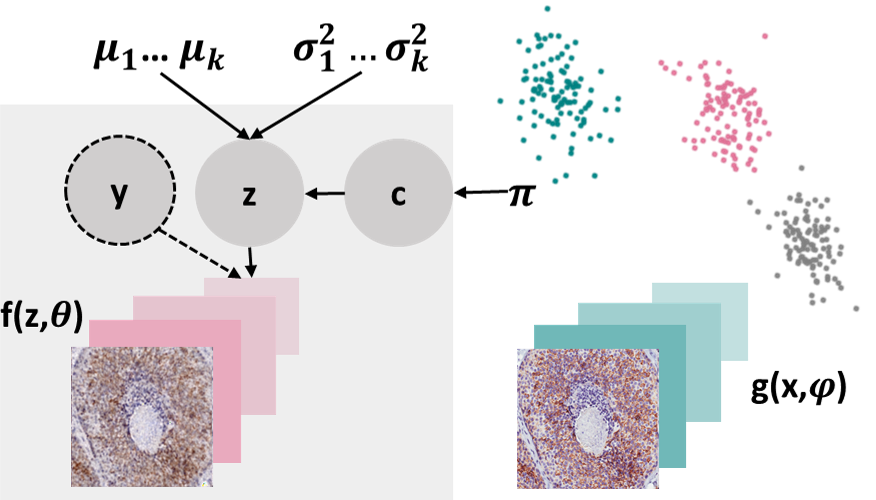
DEC Model Summary¶
Deep Embedding Clustering (DEC) [3] is a method of unsupervised learning that combines deep neural networks with clustering techniques to discover latent representations of data points. DEC involves training a deep neural network to learn a lower-dimensional representation of the data, and simultaneously optimizing a clustering loss function to group similar data points based on their embeddings.
SDCN Model Summary¶
Structural Deep Clustering Network (SDCN) is a deep neural network model that combines GCN and AE architectures for the purpose of unsupervised clustering[5].
However, original SDCN model faces significant scalability challenges that hinder its deployment in digital pathology, particularly when dealing with whole-slide digital pathology images (WSI), which are typically of gigapixel size or larger. This limitation arises from SDCN need for constructing a graph on the entire dataset and the imperative to process all data in a single batch during training. To overcome this issue, we propose batching strategy to the SDCN training process and introduce a novel batching approach tailored specifically for WSI data. [6]
M2YD Model Summary¶
The M2YD model is implemented as an experimental method, which combines an unsupervised VAE-based clustering neural network with simultaneous training of a neural network for a supervised classification task. At the current stage, the method/model is purely experimental (with limited validation), and thus not recommended for practical use, unless you know exactly what you are doing.
AE+K-means Model¶
A two-stage approach where K-means clustering (a conventional clustering algorithm) is applied to the embedding space of a trained AE. This primarily serves as a baseline for performance comparisons.
References¶
[1] Jiang, Zhuxi, et al. “Variational deep embedding: An unsupervised and generative approach to clustering.” IJCAI 2017. (https://arxiv.org/abs/1611.05148)
[2] Kingma, Welling. “Auto-encoding variational bayes.” ICLR 2013. (https://arxiv.org/abs/1312.6114)
[3] Xie, Girshick, Farhadi. “Unsupervised Deep Embedding for Clustering Analysis.” ICML 2016. (http://arxiv.org/abs/1511.06335)
[4] Sidulova, Sun, Gossmann. “Deep Unsupervised Clustering for Conditional Identification of Subgroups Within a Digital Pathology Image Set.” MICCAI, 2023. (https://link.springer.com/chapter/10.1007/978-3-031-43993-3_64)
[5] Bo, Deyu, et al. “Structural deep clustering network.” Proceedings of the web conference 2020. 2020. (https://doi.org/10.1145/3366423.3380214)
[6] Sidulova, Kahaki, Hagemann, Gossmann. “Contextual unsupervised deep clustering in digital pathology.” CHIL 2024.